Python 官方文档:入门教程 => 点击学习
目录1. 原理2. 代码实现3. 运行结果1. 原理 对于DNA序列,一阶马尔科夫链可以理解为当前碱基的类型仅取决于上一位碱基类型。如图1所示,一条序列的开端(由B开始)可能是A、T
对于DNA序列,一阶马尔科夫链可以理解为当前碱基的类型仅取决于上一位碱基类型。如图1所示,一条序列的开端(由B开始)可能是A、T、G、C四种碱基(且可能性相同,均为0.25),若序列的某一位是A,则下一位碱基是A、T、G、C的概率分别为0.25、0.20、0.20、0.20,下一位无碱基(即序列结束,状态为E)的概率为0.15。
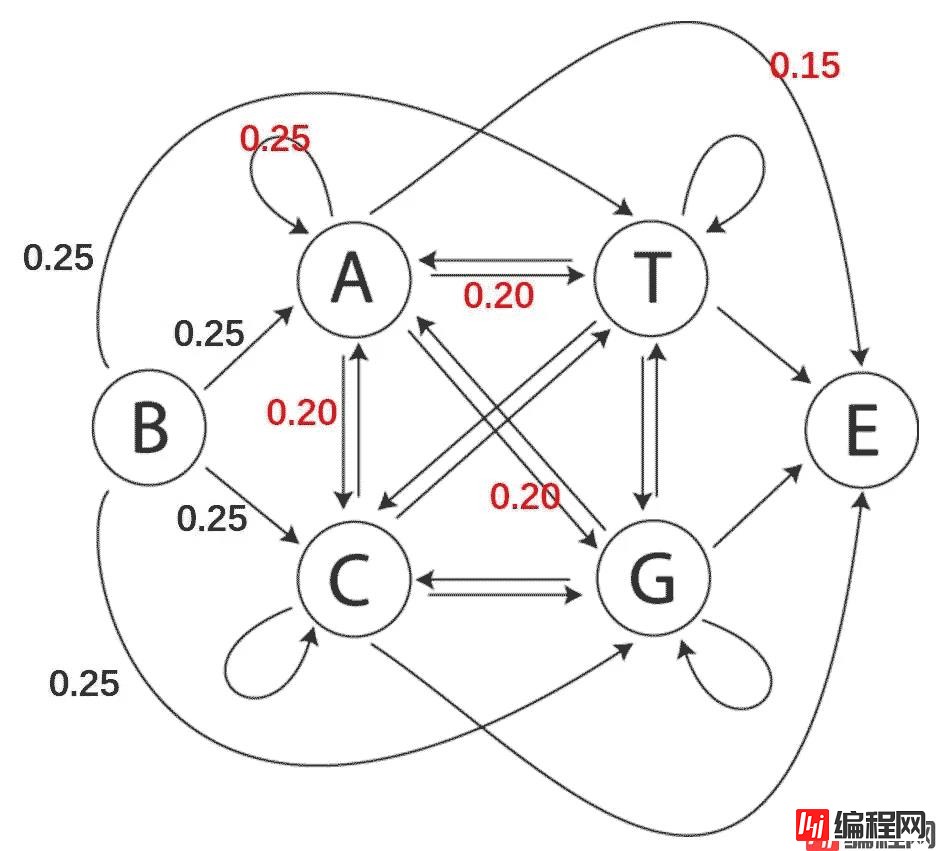
图1 DNA序列的一阶马尔科夫链
以下代码运行于Jupyter Notebook (python 3.7);代码功能是随机生成一定数量的DNA序列,统计序列长度并绘制分布图。若希望显示随机生成的序列,将代码# print(''.join(Seq))前的#删除即可。
import numpy
import random
import seaborn as sns
import matplotlib.pyplot as plt
# 状态空间
states = ["A","G","C","T","E"]
# 可能的事件序列
transitionName = [["AA","AG","AC","AT","AE"],
["GA","GG","GC","GT","GE"],
["CA","CG","CC","CT","CE"],
["TA","TG","TC","TT","TE"],]
# 概率矩阵(转移矩阵)
transitionMatrix = [[0.25,0.20,0.20,0.20,0.15],
[0.20,0.25,0.20,0.20,0.15],
[0.20,0.20,0.25,0.20,0.15],
[0.20,0.20,0.20,0.25,0.15]]
def RandomDNAs(Num):
max_len = 0
i = 0
Seq = [] #创建列表(Seq)用于添加碱基,以组成DNA序列
Len = [] #创建列表(Len)用于记录每条生成序列的长度
while i != Num:
Base = ["A","G","C","T"]
START = random.choice(Base) #随机从碱基中选择一个作为序列的起始碱基
Seq.append(START) #将起始碱基添加至Seq中
while START != "E":
if START == "A":
change = numpy.random.choice(transitionName[0],p=transitionMatrix[0])
#以transitionMatrix矩阵第一行的概率分布随机抽取transitionName第一行包含的事件
if change == "AA":
START = "A" #如果转移状态是AA(即A碱基接下来的碱基是A,则将起始碱基设为A)
elif change == "AG":
START = "G"
elif change == "AC":
START = "C"
elif change == "AT":
START = "T"
elif change == "AE":
START = "E"
elif START == "G":
change = numpy.random.choice(transitionName[1],p=transitionMatrix[1])
if change == "GA":
START = "A"
elif change == "GG":
START = "G"
elif change == "GC":
START = "C"
elif change == "GT":
START = "T"
elif change == "GE":
START = "E"
elif START == "C":
change = numpy.random.choice(transitionName[2],p=transitionMatrix[2])
if change == "CA":
START = "A"
elif change == "CG":
START = "G"
elif change == "CC":
START = "C"
elif change == "CT":
START = "T"
elif change == "CE":
START = "E"
elif START == "T":
change = numpy.random.choice(transitionName[3],p=transitionMatrix[3])
if change == "TA":
START = "A"
elif change == "TG":
START = "G"
elif change == "TC":
START = "C"
elif change == "TT":
START = "T"
elif change == "TE":
START = "E"
if START != "E":
Seq.append(START) #如果状态转移后不为End(E),则将转移后的碱基加到Seq序列中
i += 1
Len.append(len(Seq))
if len(Seq) > max_len:
max_len = len(Seq)
#print(''.join(Seq))
Seq.clear()
plt.hist(numpy.array(Len), bins=max_len, edgecolor="white")
# 显示横轴标签
plt.xlabel("DNA Sequence Length")
# 显示纵轴标签
plt.ylabel("Frequency")
# 显示图标题
plt.title("Histogram of frequency distribution of DNA sequence length")
plt.show()
print("DNA序列的最大长度为:",max_len)
print("DNA序列长度的众数为:",max(Len, key=Len.count))
%matplotlib notebook #若未使用Jupyter Notebook,此句不需要
RandomDNAs(1000) #1000表示随机生成1000条序列
从以下4个序列长度分布统计可以看到,随着随机生成的序列数量增多,序列长度分布愈发集中,且长度为1bp的序列占比最多且逐渐增加。
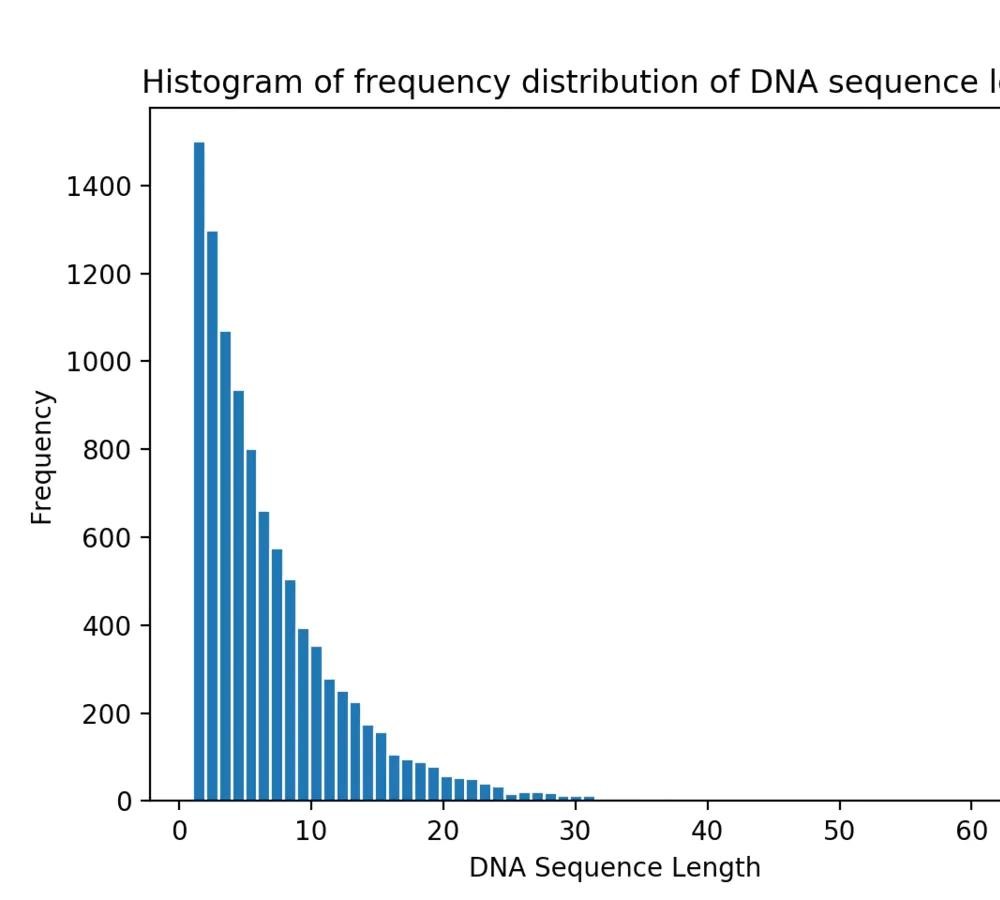
图2 10,000条DNA序列的序列长度分布统计
10,000条DNA序列的序列中
DNA序列的最大长度为: 65
DNA序列长度的众数为: 1
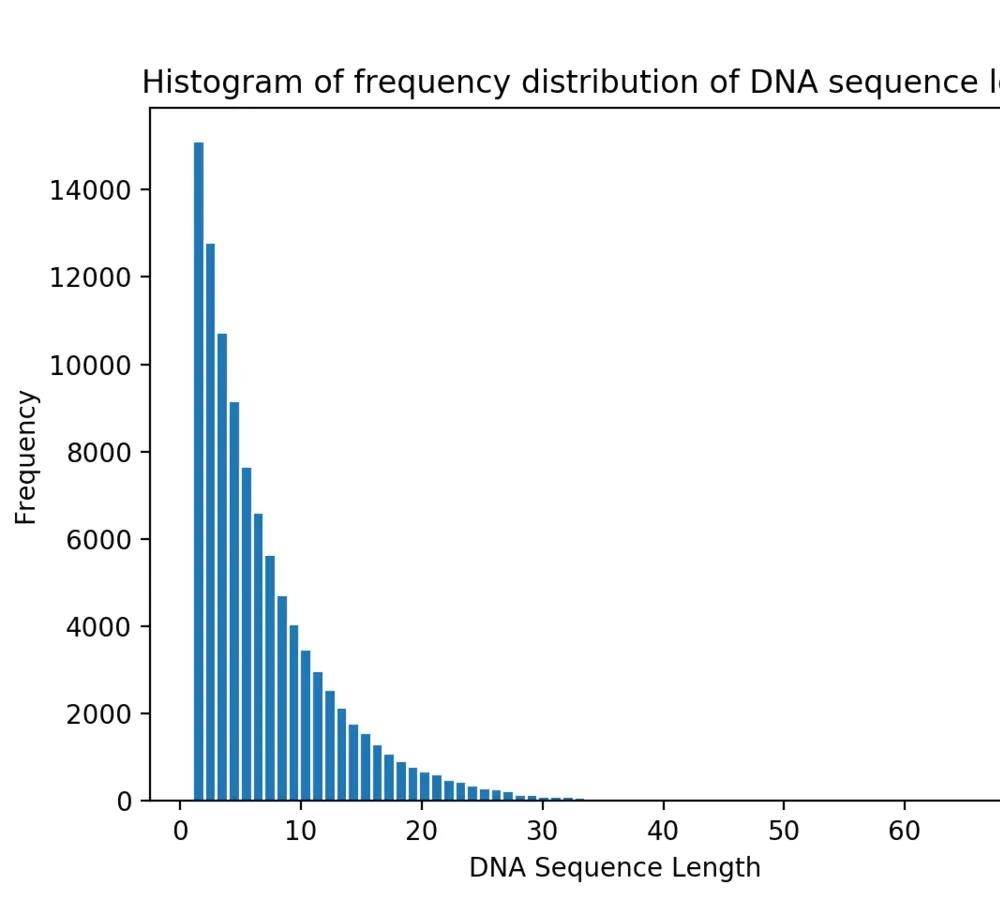
图3 100,000条DNA序列的序列长度分布统计
100,000条DNA序列的序列中
DNA序列的最大长度为: 71
DNA序列长度的众数为: 1
以上就是Python实现一阶马尔科夫链生成随机DNA序列的详细内容,更多关于Python一阶马尔科夫DNA序列的资料请关注编程网其它相关文章!
--结束END--
本文标题: Python一阶马尔科夫链生成随机DNA序列实现示例
本文链接: https://lsjlt.com/news/119060.html(转载时请注明来源链接)
有问题或投稿请发送至: 邮箱/279061341@qq.com QQ/279061341
2024-03-01
2024-03-01
2024-03-01
2024-02-29
2024-02-29
2024-02-29
2024-02-29
2024-02-29
2024-02-29
2024-02-29
回答
回答
回答
回答
回答
回答
回答
回答
回答
回答
0